Abstract
Background
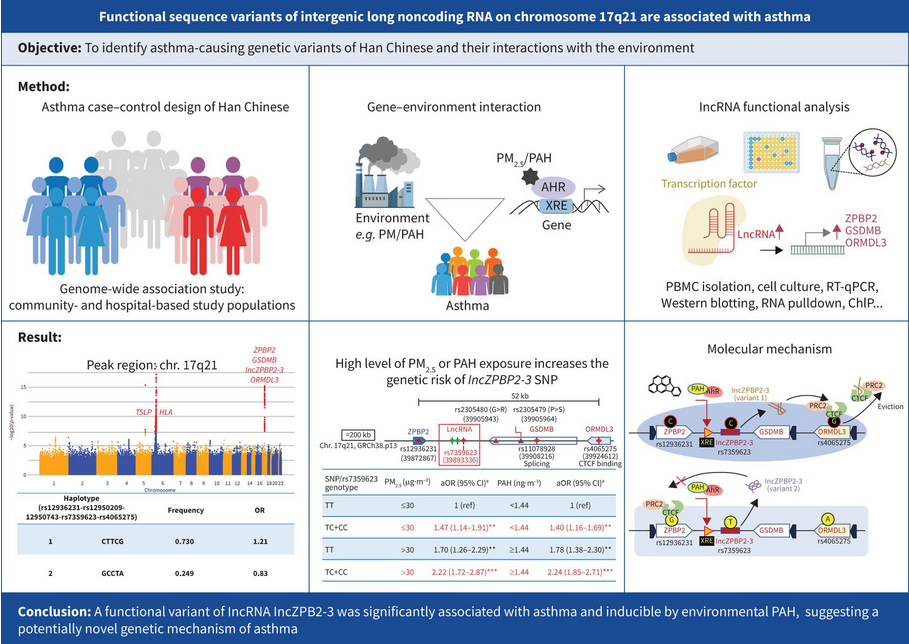
The genetic and molecular basis of asthma remains unclear and its gene–environment interaction is still enigmatic. In the present study, we aimed to identify asthma-causing genetic variants and their interactions with the environment.
Methods
We performed case–control genome-wide association studies on individuals of Han Chinese descent from the Taiwan Biobank (n=4877 cases, n=98 218 controls) to identify asthma susceptibility loci, validated in a hospital-based population of subjects (n=2595). The 10–15-year exposures to cumulative ambient particulate matter with a diameter of <2.5 μm (PM2.5) and polycyclic aromatic hydrocarbons (PAHs) were assessed for gene–environment relationships. The function of the newly identified long noncoding RNA lncZPBP2-3 and its interactions with PM2.5 and PAH exposure were analysed using RNA immunoprecipitation, RNA pulldown, reverse transcription quantitative PCR and Western blotting.
Results
Chromosome 17q12-21 was found to be a significant risk region, encompassing variants of lncZPBP2-3 and its neighbouring genes, which interacted with increasing exposure to PM2.5 and adsorbed PAHs. The expression of lncZPBP2-3 was elevated, correlating with the expression of its neighbouring genes, in the peripheral blood of patients with asthma compared to that in controls. Unlike non-risk lncZPBP2-3, the risk variant of lncZPBP2-3 disrupted transcriptional suppression of the risk locus via its interaction with the transcription insulator CCCTC-binding factor, concomitant with the higher expression levels of neighbouring genes in individuals with the risk genotype.
Conclusion
A functional variant of lncRNA, lncZPBP2-3, was significantly associated with asthma and is inducible by environmental PAH, suggesting a potentially novel genetic and molecular mechanism of asthma.